Views
From TheGPMWiki
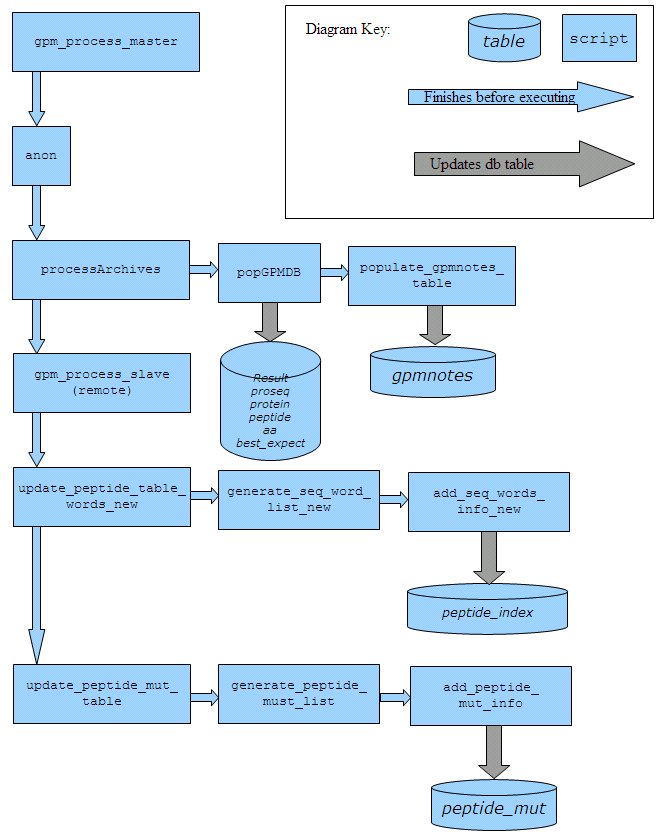
In the workflow below, script names are in bold and database table names are in italic.
- calls function set_lock
- filesystem change: makes a lock file
- calls system "gzip"
- filesystem change: unzips all xml files in archive directory
- calls script anon.pl
- filesystem change: removes some data from xml files in archive directory
- calls function archive_files
- filesystem change: copies files from archive directory to their proper subdirectory.
- calls function gzip_files
- filesystem change: gzips all xml files in archive directory.
- calls function ftp_archive
- calls system "ftp" on file list for each client server.
- calls function process_archives
- calls script processArchives.pl
- for each GPM file in archive directory
- calls script popGPMDB.pl
- parses the xml file for data to add to the gpmdb tables:
- tables inserted into: result, proseq, protein, peptide, aa
- tables updated/inserted into: best_expect
- calls script populate_gpmnotes_table.pl
- parses the xml file for data to add to enspmapdb
- tables inserted into or updated: gpmnotes
- parses the xml file for data to add to enspmapdb
- parses the xml file for data to add to the gpmdb tables:
- calls script processArchives.pl
- calls function ftp_statistics
- calls system “ftp” to send statistics file to each client server
- filesystem change: creates and deletes ftp script
- calls system “ftp” to send statistics file to each client server
- calls function update_slaves
- uses LWP to trigger script gpm_process_slave.pl on each client server
- calls function update_words
- uses LWP to trigger script gpm_process_slave.pl on each client server
- calls script update_peptide_words_table_new.pl
- script for updating the peptide_words_index table for faster tag searches
- calls script generate_seq_word_list_new.pl
- checks for peptide sequences not yet added to peptide_words table, and creates a new file if oldest found file is out of date.
- Filesystem change: may add a file to the log directory containing nonredundant list of peptide sequences added since last update.
- calls script add_seq_words_info_new.pl
- queries for pepids newer that previous max pepid to add to peptide_words table
- tables inserted into: peptide_word_index
- queries for pepids newer that previous max pepid to add to peptide_words table
- calls function update_peptide_mutations
- script for updating the peptide_mut table.
- calls script update_peptide_mut_table.pl
- calls script generate_peptide_mut_list.pl
- generates list of recently-added files, peptides, proteins and spectrum id that contain mutations, if any.
- Filesystem change: may add a file to the pep_mut directory that contains location information of peptides that contain a mutation.
- calls script add_peptide_mut_info.pl
- updates the database with mutation information in the just-added files.
- Tables inserted into: peptide_mut
- updates the database with mutation information in the just-added files.
- calls script generate_peptide_mut_list.pl
- calls function cleanup_files
- cleans up the archive directory
- calls system "del"
- filesystem change: deletes the gzip files in the base archive directory.
- calls function remove_lock
- filesystem change: deletes the lock file